02-Preprocessing patches for burned area mapping¶
In this step patches downloaded from Google Earth Engine (GEE) will be
preprocessed as input for applying the U-net architecture in order to
map burned areas. We will kick off by reading TFRecod format from Google
Drive, and finish with some patches visualizations using the scikit-eo
Python package.
01. Libraries to be installed:¶
1 | |
02. Libraries to be used:¶
1 2 3 4 5 6 7 8 9 | |
Count the number of cores in a computer:
1 2 | |
Connecting to Google Drive: This step is very importante considering that the downloaded patches are inside Google Drive.
1 2 | |
03. Reading patches dowloaded from Earth Engine¶
1 2 3 4 5 6 7 8 9 | |
04. Display metadata for patches¶
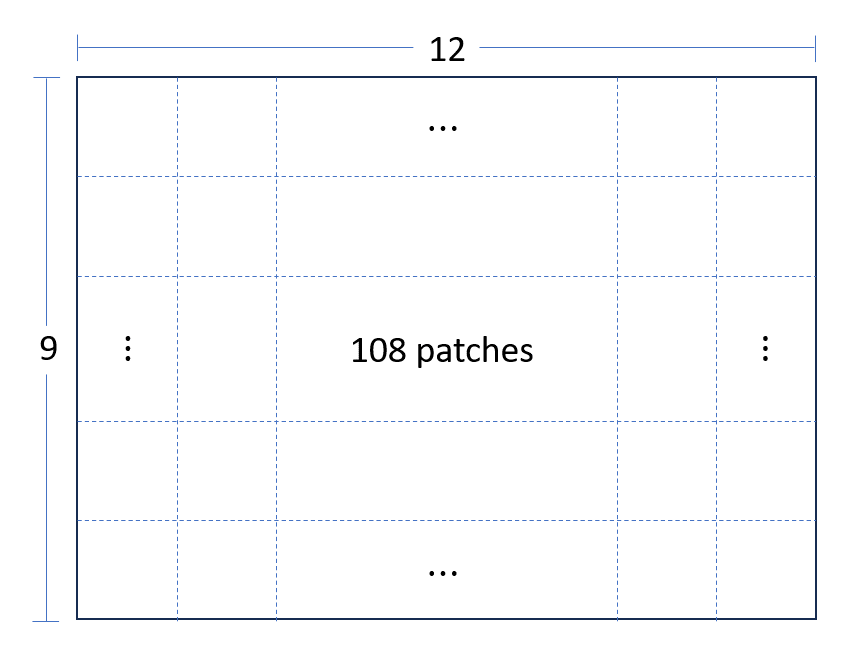
Patch dimensions (pixels per rows, cols), patches per row and total patches are crucial for the unpatched process.
1 2 3 4 5 6 7 | |
Number of patches per row and col is display in the following image:
Getting patch dimensions an total patches:
1 2 3 4 5 6 | |
05. Define the structure of your data¶
For parsing the exported TFRecord files, featuresDict is a mapping
between feature names (recall that featureNames contains the band and
label names) and float32 tf.io.FixedLenFeature objects. This mapping
is necessary for telling TensorFlow how to read data in a TFRecord file
into tensors. Specifically, all numeric data exported from Earth Engine
is exported as float32.
Note: features in the TensorFlow context (i.e.
tf.train.Feature) are not to be confused with Earth Engine features (i.e.ee.Feature), where the former is a protocol message type for serialized data input to the model and the latter is a geometry-based geographic data structure.
1 2 3 4 5 6 7 8 9 10 | |
06. Parse the dataset¶
Now we need to make a parsing function for the data in the TFRecord
files. The data comes in flattened 2D arrays per record and we want to
use the first part of the array for input to the model and the last
element of the array as the class label. The parsing function reads data
from a serialized Example
proto
into a dictionary in which the keys are the feature names and the values
are the tensors storing the value of the features for that example.
(These TensorFlow
docs explain
more about reading Example protos from TFRecord files).
1 2 3 4 5 6 7 8 9 10 11 12 13 | |
Note that you can make one dataset from many files by specifying a list
1 2 3 | |
07. Patches as tensors¶
In this step, patches downloaded for GEE will be saved as a tensor with
shape (a, b, c, d), 4D. Where a represents the number of patches,
b and c represent the patch dimension and d represents the number
of bands for each patch.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 | |
Then, patches can be converted to np.array. So, in total we have 108
patches with 512x512 pixels for both image and labeling data.
1 2 3 4 5 6 7 | |
Dealing with NaN values: In this step we are replacing NaN values for 0,
in case our data contain Null values. This will allow computing matrix
operations with deep learning architectures.
1 2 3 4 5 | |
08. Normalizing data¶
Machine learning algorithms are often trained with the assumption that all features contribute equally to the final prediction. However, this assumption fails when the features differ in range and unit, hence affecting their importance.
Enter normalization - a vital step in data preprocessing that ensures uniformity of the numerical magnitudes of features. This uniformity avoids the domination of features that have larger values compared to the other features or variables.
Normalization fosters stability in the optimization process, promoting faster convergence during gradient-based training. It mitigates issues related to vanishing or exploding gradients, allowing models to reach optimal solutions more efficiently. Please see this article for more detail.
Z-score normalization:
Z-score normalization (standardization) assumes a Gaussian (bell curve) distribution of the data and transforms features to have a mean (μ) of 0 and a standard deviation (σ) of 1. The formula for standardization is:
Xstandardized = (X - μ)/ σ
This technique is particularly useful when dealing with algorithms that assume normally distributed data, such as many linear models.
Z-score normalization is available in scikit-learn via StandardScaler.
1 2 3 4 5 6 7 8 | |
Labeling data only has two values 0 and 1, which 0 means unburned and 1 burned area. According to your data, labeling data could have many values.
1 2 | |
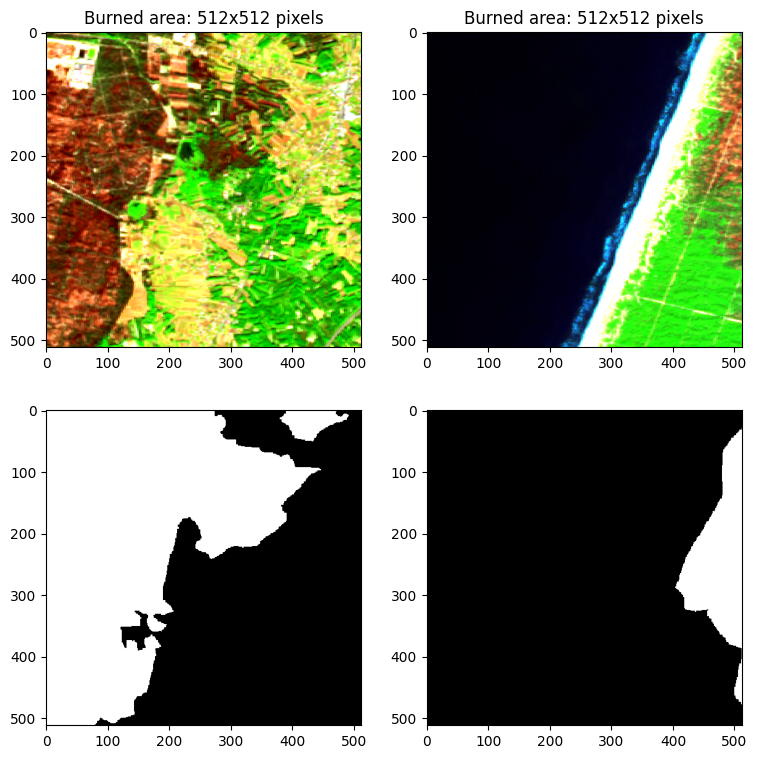
09. Visualization of patch¶
Visualizing patches and labeling data using the scikit-eo Python
package with the plotRGB function.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 | |